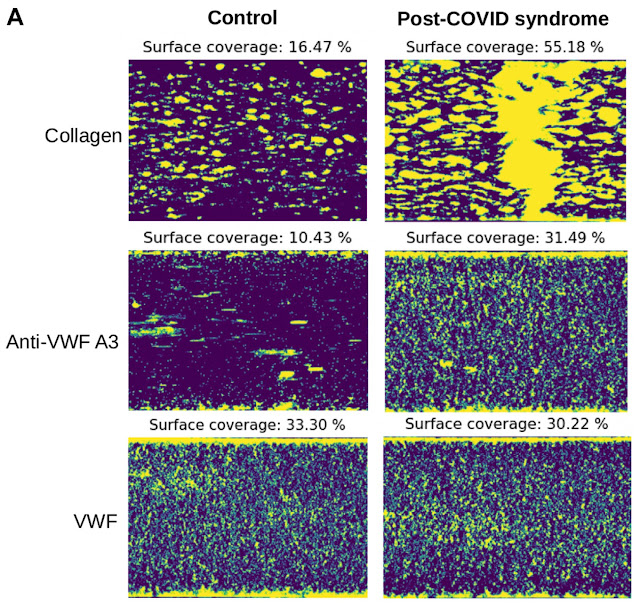
Experience with other viral infections suggests that increased autoimmunity following infection with COVID-19 might contribute to the symptoms of Post-Acute Sequelae of SARS-CoV-2 infection (PASC), more commonly known as Long COVID (LCOVID). LCOVID is characterized by the presence of autoantibodies of diverse specificities and clinical improvement is correlated with their decline. Immunoglobulin G (IgG) antibodies from fibromyalgia patients has been shown to cause pain in mice. Therefore, this group tested the effects on mice of IgG antibodies from LCOVID patients.
Most LCOVID patients’ plasma showed reduced IFN-gamma (contrast this report) and many showed elevated Glial Fibrillary Acidic Protein (GFAP, which is neuroprotective); other measured proteins were unchanged (Fig 1). Patient sera were separated into 3 groups based on neuronal damage and astroglial activation markers (Long Covid-1, LC-1), and type I IFNs (LC-2 higher IFNa2a, beta, LC-2 lower). Principal component analysis of sera proteome signatures confirmed groupings (Figs 3 & 4).
 IgG from patients with elevated neuronal proteins (LC-1) or
leukocyte activation markers (LC-3) caused sensory hypersensitivity (Fig 5,
shown; LC1 red, LC3 yellow ). They claim
that IgG from patients with higher muscle proteins also reduced locomotor
movements in mice, though the extent appears small (Fig 6). This careful study is an important guide to
such analyses.
IgG from patients with elevated neuronal proteins (LC-1) or
leukocyte activation markers (LC-3) caused sensory hypersensitivity (Fig 5,
shown; LC1 red, LC3 yellow ). They claim
that IgG from patients with higher muscle proteins also reduced locomotor
movements in mice, though the extent appears small (Fig 6). This careful study is an important guide to
such analyses.