 drj writes "Human immunodeficiency virus type 1 (HIV-1), the cause of acquired immunodeficiency syndrome (AIDS) in humans, efficiently enters Old World monkey cells but it is not reverse transcribed. This block probably protects monkeys from productive HIV-1 infection and AIDS. Stremlau and colleagues searched for an Old World monkey (rhesus) gene blocking HIV-1 infection by transferring an expression library into a susceptible human cell line (HeLa) and testing for HIV-1 resistance. They found TRIM5a, a 497 amino acid protein located in cytoplasmic bodies and named only for its structural "tripartite motif". Inhibition might result from the ubiquitin ligase activities of TRIM5a, which marks a protein for degradation but here might simply interfere with capsid disassembly. Notably, the rhesus TRIM5a does not block simian immunodeficiency virus (SIV) infection. Human TRIM5a differs in several key residues that are presumably involved in recognizing HIV-1 capsid.
drj writes "Human immunodeficiency virus type 1 (HIV-1), the cause of acquired immunodeficiency syndrome (AIDS) in humans, efficiently enters Old World monkey cells but it is not reverse transcribed. This block probably protects monkeys from productive HIV-1 infection and AIDS. Stremlau and colleagues searched for an Old World monkey (rhesus) gene blocking HIV-1 infection by transferring an expression library into a susceptible human cell line (HeLa) and testing for HIV-1 resistance. They found TRIM5a, a 497 amino acid protein located in cytoplasmic bodies and named only for its structural "tripartite motif". Inhibition might result from the ubiquitin ligase activities of TRIM5a, which marks a protein for degradation but here might simply interfere with capsid disassembly. Notably, the rhesus TRIM5a does not block simian immunodeficiency virus (SIV) infection. Human TRIM5a differs in several key residues that are presumably involved in recognizing HIV-1 capsid. Stremlau et al. Nature 427:858. February 26, 2004.
Stremlau et al. Nature 427:858. February 26, 2004.
'The cytoplasmic body component TRIM5a restricts HIV-1 infection in Old World monkeys' Matthew Stremlau, Christopher M. Owens, Michel J. Perron, Michael Kiessling, Patrick Autissier & Joseph Sodroski"
Thursday, September 7, 2006
Innate intracellular resistance to HIV-1
mRNA quality control
 drj writes "Messenger RNA (mRNA) with a premature termination codon (PTC) undergoes nonsense mediated decay (NMD). NMD is thought to protect against amino-terminal protein fragments that could be disruptive, for example, by acting as dominant negative mutants. Muscular dystrophy, Marfan syndrome, and other human diseases are caused by premature termination. Hillman and colleagues earlier determined that over one-third of alternatively-spliced human mRNAs have a PTC. Since NMD mechanisms were only recently discovered, they suspected many mRNAs in the databases might contain previously unrecognized PTCs and would have undergone NMD in vivo. Here, they aligned protein sequences from SWISS-PROT with gene sequences from Genbank to identify ~8% of entries that contain a PTC. Important biological effects could be attributed to NMD. For example, an intronic polymorphism linked to type II diabetes reduces 4 of 8 calpain-10 mRNA isoforms. These same 4 isoforms contain a PTC, suggesting NMD underlies reduced expression, causing disease. Similar analysis of CDC-like kinases and a receptor containing a death domain suggests previously unsuspected roles for NMD.
drj writes "Messenger RNA (mRNA) with a premature termination codon (PTC) undergoes nonsense mediated decay (NMD). NMD is thought to protect against amino-terminal protein fragments that could be disruptive, for example, by acting as dominant negative mutants. Muscular dystrophy, Marfan syndrome, and other human diseases are caused by premature termination. Hillman and colleagues earlier determined that over one-third of alternatively-spliced human mRNAs have a PTC. Since NMD mechanisms were only recently discovered, they suspected many mRNAs in the databases might contain previously unrecognized PTCs and would have undergone NMD in vivo. Here, they aligned protein sequences from SWISS-PROT with gene sequences from Genbank to identify ~8% of entries that contain a PTC. Important biological effects could be attributed to NMD. For example, an intronic polymorphism linked to type II diabetes reduces 4 of 8 calpain-10 mRNA isoforms. These same 4 isoforms contain a PTC, suggesting NMD underlies reduced expression, causing disease. Similar analysis of CDC-like kinases and a receptor containing a death domain suggests previously unsuspected roles for NMD. Hillman et al. Genome Biology 5:R8. January 2, 2004
Hillman et al. Genome Biology 5:R8. January 2, 2004
Background: In mammals, "premature" is a stop codon >50 nucleotides upstream of the last intron. mRNAs with PTC are flagged for degradation by the persistent splice junction protein complexes, which are normally cleared by the ribosome.
An unappreciated role for RNA surveillance R Tyler Hillman, Richard E Green and Steven E Brenner. Genome Biology 2004, 5:R8. online at genomebiology"
Wednesday, September 6, 2006
Lymphocyte migration triggered by sphingosine-1-phosphate

drj writes "Why did the lymphocyte leave the lymphoid organ? A chemical gradient was long suspected and now some details have been identified. Matloubian et al. found very few lymphocytes in the blood of mice with lymphocytes that are deficient in one particular receptor for sphingosine-1-phosphate (S1P), called S1P1. They show that developing S1P1-deficient thymocytes remain in the thymus and B cells remain in peripheral lymphoid organs, though there is only a mild alteration in the the subpopulations within these organs. Normal thymocytes but not deficient thymocytes move in response to a S1P gradient. Antigen stimulated T cells transiently reduce S1P1 expression, which is mostly restored within days, suggesting a molecular basis for transient retention in lymphoid organs. An immunosuppressant drug currently in clinical trials, FTY720, binds S1P1, leading to receptor downregulation and reduced migration. The data strongly support a compelling model, albeit one with many details yet to be determined." M. Matloubian et al. Nature 427:355-360. January 22, 2004
M. Matloubian et al. Nature 427:355-360. January 22, 2004
Enzymology - The Next Generation
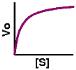 drj writes "This thought-provoking overview of current enzymology in The Scientist [free registration required] briefly introduces advances in structural studies, mechanistic theories, ribozymes, and potential commercial applications. My favorite was conceiving of the cell as a network of enzymes operating at rates determined by their (intrinsic) Km and the concentration of substrates and cofactors. Not a new idea, perhaps, but well described and evocative. The network is dramatized by these mapping of metabolic pathways and cellular and molecular processes (through ExPASy, courtesy of Roche). (originally posted on MedDot.org)
drj writes "This thought-provoking overview of current enzymology in The Scientist [free registration required] briefly introduces advances in structural studies, mechanistic theories, ribozymes, and potential commercial applications. My favorite was conceiving of the cell as a network of enzymes operating at rates determined by their (intrinsic) Km and the concentration of substrates and cofactors. Not a new idea, perhaps, but well described and evocative. The network is dramatized by these mapping of metabolic pathways and cellular and molecular processes (through ExPASy, courtesy of Roche). (originally posted on MedDot.org)
 "Enzymology's New Frontiers" M. Greener, The Scientist. 2004;Vol. 18 1:16-20 January 19, 2004"
"Enzymology's New Frontiers" M. Greener, The Scientist. 2004;Vol. 18 1:16-20 January 19, 2004"

